An Admixture Calculator for Eurasians
Genealogical testing companies use highly admixed references
In other posts, I have mentioned the problems with ancestry proportion tests from genealogical testing companies such as 23andMe, AncestryDNA, and FTDNA, and how useless the test results can be for describing the population history and admixture for individuals from various parts of Eurasia. For example, what does it mean if you are say from Pakistan, and the results from a company such as 23andMe says that you are 98% South Asian. Does it mean that all your ancestors have lived in a bubble called S Asia from time immemorial, and that you have no ancestry from other parts of the world? or say if you are from neighboring Iran, and your results say 98% Middle-Eastern, does that mean that your ancestors are from a different bubble since time immemorial, and that historically there has never been any interaction between your ancestors, and those of the ancestors of the individual from neighboring Pakistan, whose results showed 98% South Asian? Similarly, with results from a company such as AncestryDNA, showing that an individual from Iran is 80% “Caucasus” and 20% “Middle-Eastern”. The answer is of course not, because the references they use, whether they be from India, Iran, Turkey, or England, are themselves heavily admixed, and are thus “harboring” DNA from various parts of the world.
These percentages whether from AncestryDNA, 23andMe, or FTDNA should not be taken too seriously, since none use ancestral references, meaning that their references themselves are very admixed. So if a European on 23andMe scores 0.5% S Asian (which would be quite unusual), all that really means is that they are slightly more S Asian shifted than the European references they used, which could also imply some recent S Asian admixture, but that does not mean that the subject has only 0.5% TOTAL admixture from S Asia, because, if for example the European references themselves are 5% S Asian admixed (older S Asian gene flow) then the total could be 5.5% S Asian.
Results are not apples to apples
To complicate matters 1% S Asian for a European is not the same as 1% S Asian for a W Asian. Whereas the 1% S Asian for a European could translate to 6% total S Asian, if the European references have a 5% S Asian base, the 1% S Asian showing up in a result say for a Kurd or Iranian 23andMe subject could translate to a 16% TOTAL S Asian, since the Middle Eastern references, if they are Iranians, could already have a 15% S Asian base. Therefore, 1% S Asian for a northern European would not be equal to 1% S Asian for say an Iranian or Kurd.
What I am seeing in my analysis when I strip down these references down to more basic streams of ancestry from ancients such as Early Neolithic Farmers (ENF), Neolithic and Chalcolithic Iranians from the Kurdistan region (Iran N), Eastern European Hunter Gatherers (EHG), or Western European Hunter Gatherers (WHG) is that W Eurasians have much more east, north, or south Eurasian total admixture than someone would be led to believe looking over the results from genealogical testing companies.
How much total east, north, or south Eurasian admixture is the subject of this calculator, the Ancient Eurasia 20 admixture calculator, which I have been refining over the past few weeks.
Results from the program ADMIXTURE are highly volatile
Also a word about calculators created using the program ADMIXTURE4. I discovered some time ago that the results obtained using this program are highly volatile, depending on how many samples are included in the run. I should really say extremely volatile. For example, supposing we have an Italian test subject in a K=10 supervised run consisting of 10 admixture components, K1, K2, K3…….K10. I found that for example, the admixture percentage say for component K3 could vary anywhere from 0 to 20% depending on how many non-reference samples I included in the run, yes I did say non-reference samples. The reason this happens is that the allele frequencies are not entirely defined by the admixture component references, but rather, by the combination of component references, as well as the non-reference samples in the run. In other words, the admixture component allele frequencies are skewed by the non-reference samples.
I have personally created numerous admixture calculators in the past based on the program ADMIXTURE, many of which are freely available at Gedmatch.com, under project GedrosiaDNA, however, I intend to revise all of them based on my latest findings.
Generally, my experience has been that for the ADMIXTURE model the following are important criteria to keep in mind when designing a calculator:
- Independent samples. The dataset should be screened for related samples using IBD programs such as Beagle;
- There should be sufficient overlapping SNPs to resolve closely related source populations. A genotype rate of 100% for all samples is ideal, but sometimes has to be compromised due to insufficient overlapping markers between ancients and Illumina genotyped test subjects. There is a little give or take here;
- Outliers should be screened from the population sources.
One of the biggest problems with the ADMIXTURE based calculators out there, whether on Gedmatch or elsewhere, is that calculator creators use too many test samples in supervised runs. This practice seems to go back to the days when genome bloggers first started putting together calculators. The number of test samples in the run used to create the calculator ends up hugely outnumbering the reference/source populations. As previously mentioned, the test samples greatly affect the allele frequencies of the calculator component source populations, resulting essentially in a non-fully supervised test. This is because the ADMIXTURE model not only estimates Q, but also P, as a function of both the reference samples and the test samples. While the P values should remain stable regardless of the test samples, in practice the test samples change the P estimates from their actual values. This manifests itself with admixture percentage results fluctuating all over the place as the number of test samples is increased or decreased. Therefore, the number of test samples should be much fewer than the number of the reference samples and not the other way around.
Also, another point regarding ADMIXTURE in general, is that it is not informative as to direction of geneflow. An agreement in alleles can result from samples A and B sharing a common ancestor.
With regards to the slightly elevated SSA in some test samples here, it is important to remember that I don’t have a modern SW Asian component, which can cover some SSA. Also, in the interest of using a decent amount of markers, I do not have 100% marker overlap between Levant BA and the test samples, leaving some regions in the test samples genomes which are not covered by Levant BA. This of course assumes that any SSA into Eurasia or backflow from Eurasia to Africa predates Levant BA which may or may not be the case.
The Ancient Eurasia 20 admixture calclulator
I have been able to mitigate the problems described above in this calculator. Here, I only used DNA sequences from ancients and moderns that had the highest number of intersecting SNPs, with the Illumina V4 microarray, since I believe most testers use 23andMe to get genotyped. BTW, I believe that Asians are at a slight disadvantage because the Illumina is biased towards Europeans, meaning that it is not as good for picking up derived alleles in Asians, as it is for Europeans.
For my latest calculator, I use a mix of ancient and modern genomes. I reluctantly use moderns, mainly because currently, ancient DNA sequences from East, North East, and South East Asia are lacking.
This calculator is most useful for Eurasians, except perhaps if you happen to be from a population which I have used as a component reference. This calculator is not informative for sub-saharan Africans and indigenous Oceanians.
It is believed1 that most modern Europeans can almost exclusively be modeled with three primary streams of ancestry from; Western European Hunter Gatherers (WHG), Early European Farmers (EEF), and Eurasian steppe herders who contributed ANE (Ancient North Eurasian) and DNA from the Caucuses region . Consequently, for example, the WHG admixture percentage in the test subjects should not be interpreted as total WHG admixture in the test subject, because some to most WHG admixture is inferred by proxy by the references, who are themselves WHG admixed, who comprise the other components of the calculator. For example, some WHG would be included in the Steppe EMBA/LMBA components, because those populations also carried some WHG.
The following is a description of my admixture components:
- Altaian: Based on samples from the Altai region, this component represents the genetic contribution of the Turkic tribes as the expanded west and south into Europe and Asia over the past 1000 years.
- Scythian E/W: Based on the recently published sequences in “Ancestry and demography and descendants of Iron Age nomads of the Eurasian Steppe2″. This component is based on the combined allele frequencies of the western and eastern Scythian Iron Age samples, since there are not enough high coverage eastern or western samples to accurately source allele frequencies for separate eastern and western Scythian components. During the first millennium BCE, nomadic tribes spread over the Eurasian Steppe from the Altai Mountains over the northern Black Sea area as far as the Carpathian Basin. They also appear to have also ruled over areas of present day Iran, Kurdistan, and Afghanistan.
- Neolithic Anatolians: This is based on the 10 highest coverage 8000 year old ancient DNA sequences from Anatolia. These Anatolians introduced farming as they expanded into Europe during the neolithic.
- Neolithic & Chalcolithic Iranians (Iran N/Chl): Based on ancient DNA sequences from the Kurdistan region and vicinity in Iran, and published in “Early Neolithic genomes from the eastern Fertile Crescent3″. This component of ancestry peaks in Kurds, Iranians, Baloch, Brahui, Pashtuns, Punjabis, and some NW Indians. This component is based on the combined allele frequencies of the highest coverage Neolithic and Chalcolitic Iranian samples, as there are not enough samples to accurately source allele frequencies for separate Neolithic and Chalcolithic components. In Europeans, this component most likely represents ancestry shared by Iran N/Chl and Caucasus based ancient populations, above and beyond what was received from the Eurasian steppe herders, as Iran N/Chl are not believed to have directly contributed much to the genetics of Europeans.
- Steppe – Middle/Late Bronze Age (MLBA): Based on the allele frequencies of the 3500 year old ancient samples from the Eurasian steppe; 3 from the Andronovo culture, and 4 from the Srubnaya culture. These types of cultures are believed to have spread Indo-European languages.
- Early European Farmers: Here I used high coverage 7000 year old DNA from early European farmers; 5 from the LBK culture, 5 from Hungary, 1 from Iberia, and 1 known as Stuttgart. Individuals associated with these cultures are believed to have facilitated the spread of farming from the near east to Europe, and encountered European Hunter Gatherers who had settled Europe much earlier.
- Steppe – Early to Middle Bronze Age (EMBA): Based on allele frequencies of the highest coverage 4300-5000 year old ancient samples from the Eurasian steppe; 3 from the Yamnaya culture, and 2 from the Poltavka culture.
- Western European Hunter Gatherer (WHG): Based on 3 approximately 8000 years old ancient samples from Spain, Luxembourg, and Hungary.
- Burmese: Based on modern individuals from Burma. Most Europeans score around zero of this, however, it reaches significant levels in Kurds, Iranians, and populations further east, and represents the non-west Eurasian admixture in south and west Asians, somewhat analogous to the hypothetical Ancestral South Indian (ASI), but likely not the same.
- Amerindian-South: Based on references from the Cola and Wichi tribes of Argentina. In Europeans, this could represent shared origins between Native Americans and circumpolar peoples such as Saami.
- The remaining components are self-explanatory, and represent genetic similarity with various north and east Asian peoples.
Test results
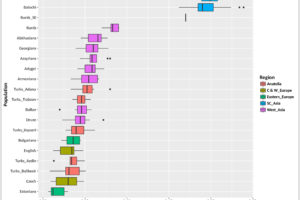
The following is a bar chart of various individuals, sorted with the highest Scythian E/W score at the bottom, and lowest at the top. It is important to remember that the Scythian E/W percentage is based on the combined allele frequencies of both eastern and western Scythians.

The following are individual results, sorted with decreasing Altaian score from left to right.


This is the FST matrix for the calculator

REFERENCES:
- Ancient human genomes suggest three ancestral populations for present-day Europeans, Lazaridis et al,Nature 513,409–413
- Ancestry and demography and descendants of Iron Age nomads of the Eurasian Steppe, Martina Unterlander et al, Nature, 2017.
- Early Neolithic genomes from the eastern Fertile Crescent, Broushaki et al, PubMed, 2016.
-
Fast model-based estimation of ancestry in unrelated individuals, D.H. Alexander, et al, Genome Research, 19:1655–1664, 2009.