In progress: How to interpret your ancestry admixture results
You have just obtained your results from a commercial ancestry result test such as 23andMe and it shows that you are 94% W Asian with 1% E Asian. Does this mean that you only have a tiny1% geneflow from some E Asian population?
You think to yourself this does not seem right especially if you happen to be one of those W Asians with E Asian physical facial characteristics. You do some online research and you come upon some blog or forum where you read that phenotype does not equal genotype. You are skeptical about this statement because you think well if phenotype does not equal genotype then why don’t Chinese people have random facial morphologies identical to Africans or NW Europeans.
You correctly conclude that the statement phenotype does not equal genotype is wrong, and that there must be some correlation between the two which causes people with “Chinese” genotypes to have “Chinese” phenotypes and not “NW European” phenotypes.
You next ponder if there is a correlation between phenotype and genotype, then why your results from 23andMe don’t show higher than 1% E Asian, considering your E Asian facial morphology.
The answer is the 1% E Asian result from 23andMe is not your total E Asian admixture. The 1% reflects E Asian admixture to the exclusion of the W Asian references used by 23andMe.
In reality those W Asian references themselves also have older E Asian admixture that has become incorporated into their genomes. The way we determine the amount of older E Asian admixture that is incorporated into the 23andMe W Asian reference genomes, is to use ancient W Asian references, such as Neolithic Near Eastern or European farmers, who predate any Iron Age or Medieval E Asian geneflow into W Asia.
If we are trying to determine total E Asian admixture in a British tester, we may choose to use Neolithic British farmers as a baseline using an ADMIXTURE based test, or a formal statistic test.
There is another reason why the E Asian admixture in a 23andMe test is underestimated. 23andMe splices your genome into 100 bp windows. The algorithm will declare a segment E Asian only if the majority of the 100 bp window contains an E Asian haplotype. In other words if you have a 40 bp E Asian haplotype which is associated with older admixture, it is disregarded in a 23andMe test.
Thus, in a 23andMe test, older E Asian admixture which is incorporated into the W Asian references is disregarded, as well as other older E Asian admixture which correlates with haplotypes less than 50 bp long.
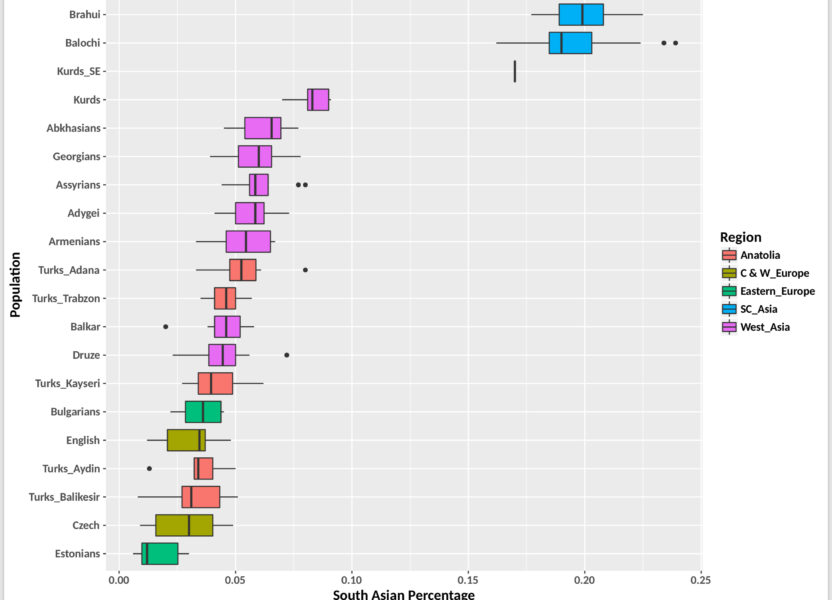
Here we attempt to quantify the TOTAL amount of minor admixture, such as E Asian in W Asian subjects such as Kurds, by using various adjacent populations, both modern and ancient as W Eurasian baselines.
We begin with 23andMe results are for 2 Kurmanji Kurd subjects, each with 4 Kurdish grandparents from the northern Iraq Kurdistan region. They are referenced as Kurd-K1 and Kurd-K3. The majority of Kurd-SE’s ancestry is from the Kurds of Iranian Balochistan and Kurds of Iraq, as well as from Iranian Baloch tribes.

Fig 1 – Kurd-K3 admixture from 23andMe

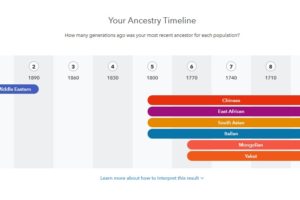
Fig 2 – Ancestry time line for Kurd-K3

Fig 3 – Kurd-K1 admixture from 23andMe

Fig 4 – Ancestry time line for Kurd-K1

Fig 5 – Kurd-SE admixture from 23andMe

Fig 6 – Ancestry time line for Kurd-SE
If you were to believe the 23andMe results, you would walk away thinking that Kurds K1 & K3 have 1% and 0.8% E Asian admixture, respectively, while Kurd-SE only has 6.8% E Asian admixture.
This of course is not the case. So where is the remainder of the E Asian admixture hidden you would ask. Well, some of it was removed by 23andMe’s “smoothing” algorithm when E Asian haplotypes smaller than 50 bp in length, which are usually associated with older admixture were tossed out.
The remainder of the E Asian admixture is incorporated into those Persian, Turkish, and Caucasian individual genetic substructure who comprise the “West Asian” references at 23andMe.
So how do we tease out this East Asian admixture which is incorporated into the Persian, Turkish, and Caucasian references. Well, we simply don’t use them as references for the West Asian component. Instead, we use references who themselves are less East Asian admixed. For example, Neolithic Near-Eastern farmers who predate all the more recent pulses of East Asian admixture into the Near East , or in a calculator based on present day references, we can use Bedouin, who themselves are less East Asian admixed than Caucasian, Persian, and Turkish references.
Here is what we get when we design a K5 ADMIXTURE based calculator (supervised mode) with the following component references from public datasets:
- South-West Asian: Negev desert Bedouin (N=19)
- West European: Basque ( N=22)
- South Asian: Paniya tribals from India (N=18)
- East Asian: Mongolians (N=22)
- Siberian: Nganasan (N=13)
Figure 7 and table 1 show the results for population averages.

Fig 7 – ADMIXTURE K-5 population averages

Table 1- ADMIXTURE K-5 population averages
The following plots show variation for individual results for the K-5 run:

Fig 8 – ADMIXTURE K-5 South Asian component individual results

Fig 9- ADMIXTURE K-5 East Asian component individual results

Fig 10- ADMIXTURE K-5 South-West Asian component individual results

Fig 11- ADMIXTURE K-5 West European component individual results
Whereas the K-5 ADMIXTURE run indicates about 9% Mongolian admixture, and about 9% South Asian admixture for Kurds-K1 and K3, their 23andMe results only show about 1% of this ancestry.
Similarly for Kurd-SE, the K-5 run shows about 21% in Mongolian admixture, and 17% South Asian admixture, 23andMe shows 6.8% East Asian and about 40% South Asian. It should be noted that the K-5 run used Paniya as South Asian references, whereas NW South Asians and Afghans are included in 23andMe’s South Asian references.
To be continued.