Impact of the Iron Age Saka and Scythians on the demography of Kurds
Overview:
The flood of recently sequenced ancient DNA has tremendously increased our understanding of demography of various Eurasian modern populations. Thanks to the recently sequenced genomes in Damgaard et al, Narasimhan et al, Olaide et al, Lazaridis et al, and Mathieson et al, and the suite of tools within Admixtools, detailed in Patterson et al we are finally able to uncover the shroud surrounding the ethnogenesis of a relatively poorly understood population; the Kurds.
Utilizing multiple lines of evidence, we were able to determine with a high degree of certainty the ancestral populations which hybridized to form present day Kurds. The analysis was performed via carefully designed tests using formal statistical methods, utilizing the various suite of tools, such as dstats and qpAdm, contained in the ADMIXTOOLS software package at Reich Lab.
Although it has been relatively easy to determine that the 5000 year old chalcolithic Zagrosian populations from Iran (Seh Gabi and Haji Firuz) formed the majority genetic input of present day Kurds, the remaining genetic input which relates to admixture from various Eurasian steppe and Central Asian populations was not as easy to ascertain until now.
The confounding issue has been that various genetic tests such as ADMIXTURE, PCA, and IBS, test for allele sharing and total shared genetic drift, but since Eurasian steppe populations, and Kurds are both substantially West Asian derived, the agreement in alleles or shared genetic drift may simply be a result of shared distant ancestry between Kurds and various steppe populations, and not necessarily due to introgression from the Eurasian steppe.
Here we are able to determine the character of this Eurasian steppe and Central Asia admixture layer on top of their 8000 year old Zagrosian farmer core. Somewhat surprisingly, this introgression is not primarily from 3500-4000 year old Middle or Late Bronze age (MLBA) steppe cultures related to Sintashta, Srubna, or Andronovo, but rather from 2000-2500 year old Iron Age (IA) populations related to Scythians and Saka, and from Middle Age (~800 year old) Turkic populations related to Kipchaks and Karakhanids.

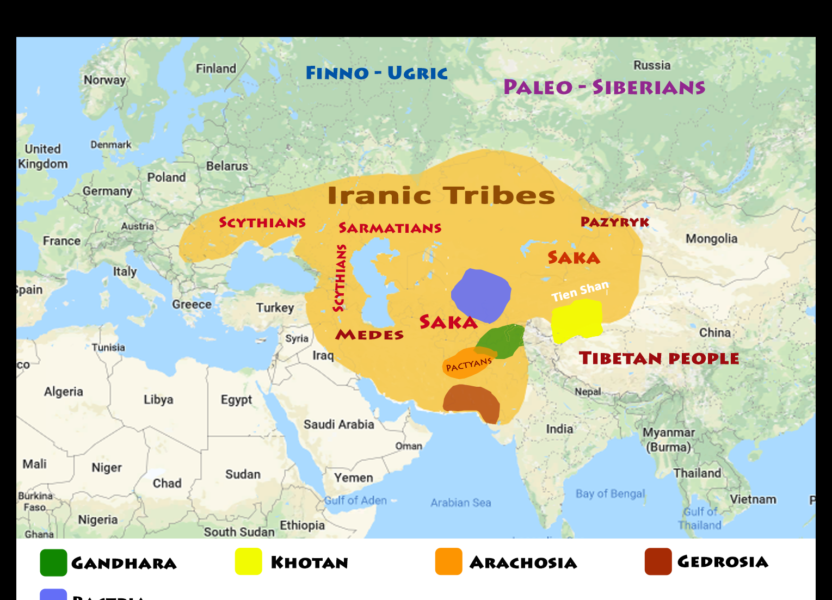
Fig 1 – Eurasian tribes of the Iron Age
Methodology:
The 8000 year old Iranian Chalcolithic samples used here were featured in previously published papers, and consist of 5 samples from the Seh Gabi area, which is on the outskirts of Kermanshah, which is within the Kurdistan region of Iran (West-Central Iran). They also consist of 5 samples from the Haji Firuz area, which is also located in the Kurdistan region of Iran (NW Iran).
The Saka and Hun Iron Age samples are also from previously published papers, and are from the Tien Shan region, which is located near the borders of Kyrgystan, Kazakhstan, and China.
The software qpAdm which is included in ADMIXTOOLS at Reich Lab, and is extensively utilized to obtain admixture proportions. Since there is some confusion regarding the code, some details are provided here based on prior experience with the software. QpAdm is a formal statistical test which uses the Likelihood Ratio Test (LRT) to test null hypotheses against alternate hypotheses.
The null hypothesis used consists of modeling a target as a combination of source populations which are analyzed against outgroups, which are phylogenetically more distant from the target. Thus the test is more informative to introgression instead of genomic similarity due to general shared drift.
For example, if a target is modeled with 3 ancestry streams as T = ∝ PopA + β PopB + γ PopC, the program calculates the p-value associated with the model. The p-value is a probability of the null hypothesis being true, ie the probability of there being no (null) difference between the observed admixture proportions and the expected admixture proportions, ∝, β, & γ. Thus a p-value of 0.5 would indicate that there is a 50% probability that the mixture model for T or the null hypothesis is true. By contrast a p-value of 0.01 would indicate that there is only a 1% probability that the mixture model is correct, in which case we can reject it based on a threshold of p>0.05.
Therefore, higher p-values in qpAdm indicate less discrepancy between the expected and observed values, which in turn confers greater confidence in the mixture model for the test subject. This is also referred to as a better fit.
Generally, the analysis is performed as follows:
- We start with knowns and solve for unknowns. Here the known core ancestral populations for Kurds are Chalcolithic Iranians, such as the Haji-Firuz genomes;
- We then test using the following dstats; D [ Mbuti/Chimp, steppe ; Iran-Chl, Kurds ] to determine which steppe populations Kurds share the most drift with to the exclusion of Iran-Chl;
- Test f3 [ Kurds; Iran, steppe ] to determine which steppe – Iran-Chl combinations produce the strongest signals of admixture for;
- Finally, using qpAdm, we determine high confidence mixture models for Kurds, using p-values, using a core Zagrosian Chalcolithic population along with a steppe population such as Sintashta-MLBA, Iron Age Saka or similar using various outgroups. The qpAdm methodology also provides a formal test of whether the model of the Test population as a mixture of 2 or 3 source streams that are clades with West Eurasians and East Eurasians, is a fit to the data, with all outgroups which are phylogenetically more distant.
The qpAdm test gives a p–value which is between 0 and 1. Generally, the p-value threshold is set at 0.05. P-values > 0.05 indicates that we can’t reject the mixture model, with higher p-values indicating greater confidence in the models.
QpAdm gives us a p-value for whether the matrix is rank=1 ( consistent with 2 streams of ancestry), or rank=2 ( consistent with 3 streams of ancestry), and an estimate of the proportion ? of ancestry that is a clade with various Steppe-MLBA or Steppe-IA populations.
With the qpAdm test, very low p-values could also indicate that the relationship of the test population to the outgroups may be more complex, and that the outgroups may be interact with the test population outside the source populations. In this case outgroups could be dropped or changed one at a time to determine whether a passing p-value can be obtained. The disadvantage of dropping an outgroup is that we are removing potentially phylogenetic useful branching points, which may result in an increase in standard errors.
Larger standard errors can indicate that we are still missing aDNA which carries important phylogenetic information not present in the current outgroups used.
Steppe-IA versus Steppe-MLBA geneflow into West Asia
Here we show that for Kurds, there is substantially greater introgression from Iron Age Central Asian populations such as Scythians, Saka, and Huns, than from MLBA steppe populations such as Andronovo, Srubna, and Sintashta.
A couple of big hurdles in academia in determining whether Steppe-MLBA or Steppe-IA had the biggest impact on the demography of Kurds have been:
- A shortage of appropriate Central Asian aDNA sources to date;
- The wrong questions have been addressed in analyses.
With regards to (1) above, we now have a decent amount of aDNA from Central Asia to make more accurate calculations thanks to the aDNA published in Damgaard et al, 2018, and Narasimhan et al, 2018,
With regards to (2) above, much of the research seeks to assess steppe ancestry in Kurds via ADMIXTURE, PCA, and IBS. The problem with doing so is that only total accumulated mutations, and total shared drift between West Asian and Steppe populations, since Out-of-Africa (OOA) is addressed. Since Steppe-MLBA is more West Eurasian than Steppe-IA, these types of analyses will overestimate Steppe-MLBA, not necessarily because of greater introgression into West Asians such as Kurds, but rather because they are substantially more derived from the same ancestral core populations that contributed to them, ie Iran-N, CHG, and Antatolia-N related.
The proper question we should seek to answer with analyses would be whether modern West Asians such as Kurds are differentially more Steppe-IA or Steppe-MLBA shifted POST Chalcolithic Iran.
Modern Kurds compared with their Zagrosian Chalcolithic Ancestors
One of the most important questions to answer is what demographic events have taken place in the Kurdistan area that differentiate modern Kurds from their Zagrosian Chalcolithic ancestors, which are here represented by the genomes from the Seh Gabi and Haji Firuz areas, which are within the Kurdistan region of Iran.
Here dstats of the form D [ Kurds, Haji-Firuz; Steppe, Mbuti ] can be used to compare various Kurdish samples from the Iraq/Iran area with Haji-Firuz for shared drift with steppe samples post since Haji-Firuz-Chl. Kurds C1-C3 are Kurmanji Kurd samples from northern Iraq, and Kurds F1-F7 are Feyli Kurd samples from around the Iraq/Iran border.
Here we show that the most important demographic events affecting Kurds in the Kurdistan region since the Chalcolithic involve introgression of smaller amounts of DNA from Middle to Late Bronze Age cultures related to Sintashta and Andronovo, followed by introgression of larger amounts of DNA related to Saka and Hun steppe nomads during the Iron Age, and related to Turkic Medieval populations such as Kipchaks and Karakhanids. It is the latter mechanism that explains the significant increase in East Eurasian admixture in present day Kurds when compared to their Iranian Chalcolithic forefathers.
This increase in East Eurasian admixture for present day Kurds is easily observed with dstats of the form D [Kurds, Haji-Firuz; Steppe, Mbuti ]. The results are shown in figures 2 – 4.
With regards to Steppe-MLBA, dstats indicate a dilution of this type of ancestry post Haji-Firuz, especially with some of the Feyli Kurd samples.

Fig 2- Dstats of the form D [Kurds, Haji-Firuz; Steppe, Mbuti ] showing the biggest change since Haji-Firuz-Chl is E Asian geneflow into modern Kurds

Fig 3 – Dstats of the form D [Kurds, Haji-Firuz; Steppe, Mbuti ] showing the biggest change since Haji-Firuz-Chl is E Asian geneflow into modern Kurds

Fig 4 – Dstats of the form D [Kurds, Haji-Firuz; Steppe, Mbuti ] showing the biggest change since Haji-Firuz-Chl is E Asian geneflow into modern Kurds
The dstats shown in figures 2-4 don’t support introgression of Steppe-MLBA such as Sintashta and Andronovo into the Kurdistan region, post Haji-Firuz-Chl, however, this is also further investigated via the qpAdm method.
To determine whether modern Kurds have a larger layer of Steppe-IA, or a larger layer of Steppe-MLBA layer on top of their Iranian Chalcolithic core, dstats of the form D [ Mbuti, Steppe-IA; Haji-Firuz-Chl , Kurds ] vs. D [ Mbuti, Steppe-MLBA; Haji-Firuz-Chl , Kurds ] are used to shed light on whether Kurds differentially share greater genetic drift with Steppe-MLBA populations, or with Iron Age Saka and Huns, to the exclusion of their Haji-Firuz-Chl core. Thus it is helpful to compare the drift path lengths of Kurds & IA Tien Shan Huns against Kurds & MLBA steppe populations, subsequent to Chalcolithic Iran. In figure 5 we see that every Kurdish test sample is differentially more net Tien Shan Hun shifted than Sintashta-MLBA shifted, to the exclusion of Haji-Firuz-Chl.

Fig 5 – Differential shared genetic drift comparison of Iron Age Tien Shan Huns vs MLBA Sintashta to the exclusion of Haji-Firuz-Chl for Kurds
The strategy we use with the qpAdm method is to let the program pick admixture proportions based on 3 source populations (pleft), using outgroups (pright) which are phylogenetically more distant from the Kurdish test subjects, than the 3 source populations. This will also enable us to reject models which are not a fit to the data. The combinations of sources we use are;
- Haji-Firuz-Chl, Saka-TienShan, Sintashta-MLBA
- Haji-Firuz-Chl, proto-Turkic XiongNu_WE, Sintashta-MLBA
We highlight higher confidence models based on p-values. We first check to see fits for models consisting of 3 ancestry streams; Chalcolithic Iranians , Saka, Sintashta-MLBA, using outgroups (pright) which are phylogenetically more distant from the Kurdish test subjects, than the 3 ancestry sources (pleft). This will also enable us to reject models which are not a fit to the data.
The following outgroups (pright) were used throughout, which produced the highest confidence qpAdm models for Kurds:
Mbuti
ShamankaEN
Karitiana
Ust_Ishim
WHG
West_Siberia_N
Anatolia_N
Ganj_Dareh_N
Onge
EHG
Table 1 shows that for 5 of the 9 tested Kurd samples, the 3-way models with Sintashta-MLBA included were infeasible. However,the models become feasible for all Kurdish samples when Sintashta is dropped, and Kurds are modeled as a 2-way combination of [Haji-Firuz-Chl – Saka] or [Haji-Firuz-Chl – proto-Turkic Iron Age XiongNu].

Table 1 – QpAdm 3-ancestry model for Kurds. Models with Sintashta-MLBA as a source are feasible for only about half of the Kurdish samples.
The p-values for the 2 ancestry stream models consisting of Haji-Firuz-Chl – Saka, or Haji-Firuz-Chl XiongNu-IA, ranged between 0.19 and 0.97 for the Kurdish samples. Thus, unlike the aforementioned models with Sintashta-MLBA included, we were not able to reject any of those models for the Kurdish samples. Standard errors were relatively low, and in the single digits (tables 2 and 3)

Table 2- QpAdm 2-ancestry model consisting of Haji-Firuz + Saka-TienShan

Fig 6 – QpAdm 2-ancestry model for Kurds consisting of Haji-Firuz + Saka-TienShan

Table 3 – QpAdm 2-ancestry models for Kurds consisting of Haji-Firuz + XiongNu

Fig 7 – QpAdm 2-ancestry model consisting of Haji-Firuz + XiongNu
Unlike the previous 2-ancestry models consisting of Haji-Firuz and Saka, or Haji-Firuz and XiongNu, which we could not reject, we can reject a 2-ancestry model consisting of Haji-Firuz and Sintashta-MLBA, as p-values are significantly below our threshold of 0.05 for all but one of the Kurdish samples (Table 4).

Table 4 – QpAdm 2-ancestry model consisting of Haji-Firuz-Chl & Sintashta-MLBA can be rejected for all the Kurdish samples
Although the 2 ancestry stream models consisting of Haji-Firuz-Chl and Sintashta-MLBA are infeasible and can be rejected, the models with Sintashta-MLBA become feasible with the addition of Medieval Turkics such as Kipchaks (Table 5)

Table 5 – 3-ancestry source models consisting of Haji-Firuz-Chl + Sintashta-MLBA + Kipchak Medieval Turkics produce good fits.
A summary of some of the highlights of this study is as follows:
- There is a very high degree of genetic continuity in the Kurdistan region over the past 8000 years, where most Kurdish samples in this study can be modeled as over 70% Haji-Firuz-Chalcolithic (HF-Chl) with a high degree of confidence.
- In 3-ancestry stream models consisting of HF-Chl, TienShan-Saka, and Sintashta-MLBA, there is evidence of less than 10% Sintashta genetic input in about half of the Kurdish samples.
- Simple 2-ancestry stream models consisting of only HF-Chl and Sintashta-MLBA can be rejected for all the Kurdish samples, as p-values are significantly below the p=0.05 threshold.
- Substantial introgression of East Eurasian DNA into present day Kurds since their 8000 year old Iranian farmer forefathers.
- There is evidence of substantial hybridization between the Chalcolithic Iranian ancestors of Kurds and Iron Age Central Asian Saka, and subsequent Turkic populations, which had a substantial impact on present day Kurdish ethnogenesis, because simple 2-ancestry stream models consisting of either HF-Chl and TienShan-Saka, or HF-Chl and proto-Turkic XiongNu-IA can not be rejected for any of the Kurdish samples, as p-values for those are significantly above the p=0.05 threshold, with the average p-value for all Kurds for the HF-Chl – Saka models being p=0.66, and the average p-value for the HF-Chl – XiongNu models being p=0.53.
- The models with Sintashta-MLBA as a source are feasible only when Turkics are added as an additional source.
UPDATE 9/17/2018
It appears likely that the early Indo-Iranians of the Bronze Age Eurasian Steppe such as the pastoralist Sintashta culture came into contact with the agriculturalists of the Bactria Margiana Archaeological Complex (BMAC), who themselves were derived from earlier agriculturalists who had moved east from Iran, Western Siberian Neolithic Hunter Gatherers and Ancestral South Asians ( Narasimhan 2018).
From the linguistic side there is support for this interaction because of traces of prehistoric non-Indo-European loanwords within Indo-Iranian (Damgaard 2018 Linguistic Supplement). Some of these loanwords were retained by the Iranian branch subsequent to the Iranian – Indo-Aryan split of Indo-Iranian. Such words include Hushtra camel) and kara (donkey).
This was investigated to determine whether we could pick up any evidence of this interaction between the early Indo-Iranians and the BMAC in the genetic substructure of various West Asian populations using formal stats (qpAdm) by attempting to model West Asian populations as a 4 way mix of:
- Haji-Firuz-Chl
- BMAC (Gonur-BA, Turkmenistan)
- Sintashta-MLBA
- Turkic (Medieval Kipchaks)
The following outgroups were used:
Mbuti
ShamankaEN
Karitiana
Ust_Ishim
WHG
West_Siberia_N
Anatolia_N
Ganj_Dareh_N
Onge
EHG
From the West Asian populations tested only the Kurds showed evidence of BMAC ancestry. This indicates greater Central Asian geneflow in comparison to the other populations studied, and likely explains the differential ASI shift of Kurds compared to the other West Asian populations studied. The Kurdish samples consisted of 2 Kurmanji Kurd samples from the Kurdistan region of Iraq and 4 Feyli Kurd samples.
Four populations; Georgian Jews, Bedouin, Jordanians, and Karakalpaks produced infeasible results for both the full rank model as well as all the nested models.

Table 6 – qpAdm 4 ancestry models for various West Asian populations.

Fig 8 – qpAdm 4 ancestry models for various West Asian populations. Only Kurds produced evidence of BMAC admixture indicating greater Central Asian geneflow. This likely also explains the differential Ancestral South Asian (ASI) shift of Kurds compared to the other West Asian populations studied.
The following are the individual results for the Kurdish samples studied:

Table 8 – Individual Kurdish sample results

Fig 9 – Individual Kurdish sample results
References
- The first horse herders and the impact of early Bronze Age steppe expansions into Asia, Peter de Barros Damgaard et al, 2018.
- The Genomic Formation of South and Central Asia, Vagheesh M. Narasimhan et al, 2018.
- The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods, Rui Martiniano et al, 2017.
- The Beaker Phenomenon And The Genomic Transformation Of Northwest Europe, Olaide et al, 2017.
- Genome-wide patterns of selection in 230 ancient Eurasians, Mathieson et al, 2015.
- The Genomic Formation of South and Central Asia, Narasimhan et al, 2018.
- ADMIXTOOLS, Reich Lab, https://reich.hms.harvard.edu/software.
- Ancient Admixture in Human History, Patterson et al, 2012.