In progress: A method for improving accuracy in specific shared drift tests
BACKGROUND
Over the past decade various tools have been developed for ancient DNA analysis and assessing shared drift between populations. Some programs such as STRUCTURE, ADMIXTURE are allele frequency based. Others such as Reich Lab’s ADMIXTOOLS use both allele frequencies as well as direct allele comparisons. Others yet such as IBS and IBD compared genomes for allele matches, and shared haplotypes. However, shared drift calculation accuracy due to relatively recent gene flow between an ancient population and a contemporary one is limited for the following reasons:
- The GRCh37/Hg19 Human Reference genome which was introduced in 2009, and has been used to align/map the vast majority of the aDNA sequences published to date is based on a few anonymous individuals representing a few countries and is thus not representative of human diversity. Although the donor identity ethnic group is not public, evidence based on personal experience indicates that NW Europeans and Africans are over represented. This causes a bias towards Europeans and Africans during alignment/mapping aDNA sequences to the Human Reference because some aDNA reads that fall outside of European or African variation often map to the wrong regions of the Reference genome, and sometimes don’t map at all.
Researchers and genome bloggers should be aware of some of issues outlined herein which affect accuracy of the analysis results.
We share some solutions based on personal experience which should help researchers and genome bloggers achieve higher accuracy in shared drift or admixture analysis involving ancient DNA (aDNA).
Identity-by-State (IBS)
IBS tests are useful for allele by allele comparisons of genomes, however, the results are of limited accuracy for determination of shared drift due to relatively recent geneflow from say a Bronze Age population and contemporary populations. Reasons include:
- Misalignment of regions of high variation in aDNA to the human reference genome. Alternate alleles get mapped to the wrong region of the reference genome. Based on the extensive analysis I have performed, 1/1 sites ( Homozygous variant) are problematic. Evidence for this can be seen when IBS analysis is performed on aDNA sequences from the Eurasian steppe using exclusively 1/1 positions in the genome.
OUTGROUPS BIAS SHARED DRIFT STATS
Using outgroups such as Mbuti and Chimp in comparisons involving populations such as Europeans and Asians, which have significantly different drift histories, and using SNPs ascertained in European populations, leads to inaccurate inferences that many researchers and bloggers are not aware of. Nowadays, most scientific papers published utilize the Reich Lab ADMIXTOOLS suite, which include programs such as dstats, f3s, and qpAdm to analyze shared drift between populations, and nearly always the Mbuti are used as an outgroup in the analysis.
For example, using D (Mbuti, Yamna I0231-Diploid; , Abkhazians, N Europeans) N Europeans share more drift with Yamna than Abkhazianss do. However, when Mbuti is replaced with the European Paleolithic 35,000 year old forager Goyet-Q116, the results are reversed, namely, Abkhazians share more drift with Yamna than N Europeans do. The results obtained using Mbuti, or even for that matter Chimp as an outgroup are biased for the following reasons:
- It is a well established fact that W Asians share more alleles with Mbuti and Chimp than N Europeans do using the European ascertained public panels. This can be clearly seen in the IBS runs shown in fig 1. The reason for this is two-fold;
- Relatively more recent geneflow into the W Asia to the exclusion of N Europe in the case of Mbuti;
- Ascertainment bias in the case of both Chimp and Mbuti. Polymorphic sites in the public datasets have been ascertained in W Europeans, thus as shown in the IBS runs in fig 1, W Asians share more alleles with both Mbuti and Chimp than N Europeans do, due to agreement at sites where W Asians, Mbuti, and Chimp are monomorphic to the exclusion of N Europeans.
- Although sensitivity is high, most tests in general have very low specificity. This leads to many false positive results in the comparison samples due to agreement of ancestral alleles, and not necessarily of alleles “unique” to specific populations. This is further discussed below.

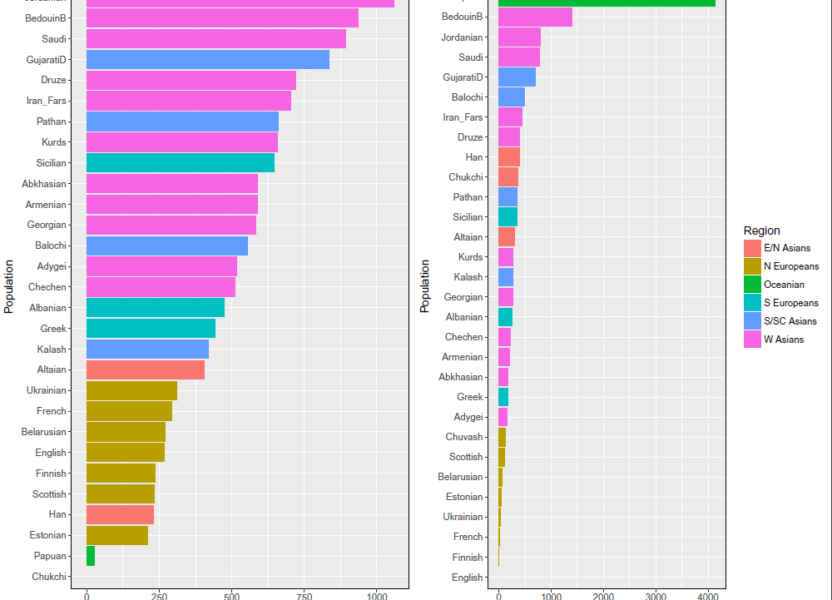
Fig 1 – Excess alleles shared by various populations over Chukchi and English, for Mbuti and Chimp, respectively. Numbers shown are population averages based on about 100K intersecting SNPs. Thus the use of Mbuti, and even Chimp as outgroups dampens the shared drift stats of W & S/SC Asians with Bronze age Eurasian steppe populations, in comparison with the stats of N Europeans with those same populations.
EXAMPLE
Let’s consider a dstat such as D (Mbuti, Yamna; W Asian, European). The idea of using an outgroup such as Mbuti to increase test specificity is good, but the results are far from accurate, if they are relied upon to determine whether W Asian or European has more geneflow from Yamna, simply because Mbuti is differentially a greater outgroup to European than W Asian. W Asians simply share more alleles with Mbuti for the aforementioned reasons, than Europeans with Mbuti. This can clearly be seen in the IBS tests shown in fig 1. If the outgroup is changed to something such as Goyet, then all of a sudden. So clearly the idea of using a single outgroup will lead to inaccurate inferences.
Thus there are 2 forces at play which act to lessen shared drift of [Asians – Steppe] when compared with [Europeans-Steppe] in D [ Asians, Europeans; Steppe, Mbuti/Chimp]. The 1st is ascertainment bias of SNPs ascertained in Europeans, and the other is African admixture into Asians.
Figure 1 is based on the folloiwng table. Here we see that neither Mbuti nor Chimp is a good outgroup for South and West Asians, because using either outgroup, South and West Asians share more alleles with Chimp and Mbuti to the exclusion of Europeans.

Table 1 – Excess allele sharing between various populations and Chimp/Mbuti to the exclusion of English based on 100K SNPs
Although neither Mbuti nor Chimp are ideal outgroups when testing West Asians such as Abkhazians, Armenians, and Kurds in D [W Asian, European; Steppe, Mbuti/Chimp], Chimp is differentially a better outgroup as (M-C) is +ve in the pink shaded column. SW Asians such as Jordanians and Bedouin are at a significant disadvantage, with Bedouin sharing 669 more alleles with Mbuti, and 1400 more alleles with Chimp to the exclusion of English. South Asians are affected similarly.
The fundamental problem with much of the analysis that is published using these tools, whether it be IBS, or something else, is that the test sensitivity is high (low false negative results), however, the test specificity is dismal ( high false positive results). Specifically:
SENSITIVITY = TP / (TP + FN); SPECIFICITY = TN / (TN + FP); PRECISION (Positive Predictive Power) = TP / (TP + FP); ACCURACY = (TP + TN) / (TP + TN + FP + FN).
where, TP = True Positives, TN = True Negatives, FN = False Negatives, and FP = False Positives.
The problem is most allele matches between Yamna and the test subjects are due to the alleles being common, and thus could have been acquired by the test subjects from a variety of sources including any EHG or CHG derived population. Thus the specificity in the test is very low because there are many false positives, ie many of the matching alleles between Yamna and the test subject likely acquired via non-Yamna. The way to increase the test specificity is to identify the small number of alleles uniquely shared by Yamna only, and not include the many alleles in common between Yamna and dozens of other populations in the test.
Although IBS is useful for assessing total shared drift between 2 populations or individuals, the results generally can’t be used to assess more specific shared drift or derived ancestry between populations, because deep common ancestry between two populations conflates any assessment of specific shared drift or derived ancestry.
Thus greater “basal” or “E Eurasian” ancestry in W Asians will dampen shared drift calculations between Yamna and W Asians as compared to Yamna and Europeans, because when deep ancestry is considered total genetic drift with Yamna is dampened for W Asians.
Here we develop and present a novel approach to assessing specific geneflow or shared drift between 2 populations by minimizing the effect of deep common ancestral contributions on the calculations.
First, an example to illustrate how deep common ancestry conflates the results and leads to wrong conclusions. Assume as shown in fig 1, we have 2 individuals, one W Asian and the other E European, and assume that they both have a Scythian parent each. In other words, individual A is 50% Scythian – 50% E European, whereas individual B is 50% Scythian-50% W Asian. Assuming that the W Asian parent is substantially Levant N and Iran N derived, whereas the E European parent is not, an analysis using total drift or IBS will show that A shares more total drift or shares more alleles with Scythians inspite of both A and B being 50% Scythian each.

Fig 1 – Individual A is 50/50 E European/Scythian. Individual B is 50/50 W Asian/Scythian
Why is this so? The reason is the E European parent shares more common distant ancestry with Scythians because he has less geneflow from clades outside the “European” clade. In other words, the E European parent has more WHG and EHG related ancestry acquired from European non-Scythian populations to the exclusion of the W Asian parent. This extra WHG and EHG related ancestry acquired by the E European parent acquired from outside of Scythians will inflate the total shared drift stats of A with Scythians inspite of A and B having exactly the same amount of Scythian ancestry, to wit, 50%.
METHODOLOGY
A solution is to designate all populations where A and B could have acquired minor/alternate alleles outside of Scythians as outgroups in an IBS analysis. In other words, any minor alleles shared between A or B and Scythian AND also occurring in outgroups such as African, ENA, WHG, EHG, CHG are not counted because those could have been acquired via other populations. Only minor alleles occurring in A or B and Scythian to the exclusion of the outgroups are counted. To solidify the results further, another Scythian sample can be introduced and only minor alleles occurring in BOTH Scythian samples and A or B can be counted.
The following results were obtained using our own custom script. Test populations, along with target populations and outgroups were pruned to intersecting SNPs only, ie a 100% overall genotype rate for the dataset. The number of positions having at least 1 minor/alternate allele in the test and target samples to the exclusion of all outgroups were counted (0/1 and 1/1 positions).
We had previously diploid genotyped the following Eurasian steppe sequences using the ATLAS pipeline which is optimized for processing aDNA sequences. This has been detailed in an earlier article on this site.:
- Scythian I0247
- Yamna Samara I0231
- Srubna I0232
- EHG I0061